The Taxonomic and Biodiversity Software Stack in R
Scott Chamberlain (@sckottie/@ropensci)
UC Berkeley - rOpenSci

scotttalks.info/tdwg17
LICENSE: CC-BY 4.0
R intro
growing in popularity
very widespread with biologists
similar to e.g., Ruby, Python, Julia
> 10K packages
a pitch for R
making biodiveristy and taxonomy software in R:
R has all parts of digital research workflow
R software means targeting users
meets scientists/etc. where they are, which:
facilitates reproducible science
facilitates open science
rOpenSci?
non-profit
make open-source software for R
large community developers/users
target science use cases
we often play role of communicating btw. scientists/data provders
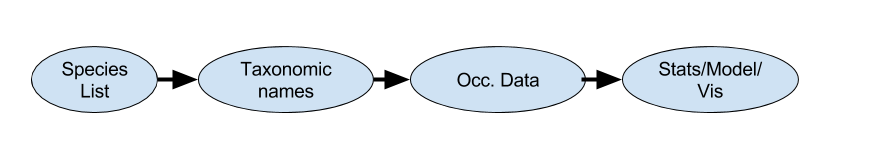
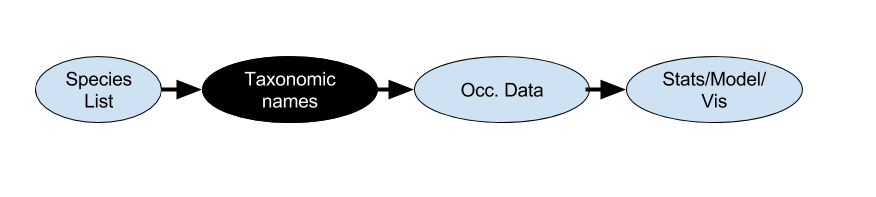
the R taxonomy landscape
R packages
* not rOpenSci
ritis - ITIS API client
taxa - taxonomic classes
taxize - one stop shop for data
taxizedb - work w/ database dumps
wikitaxa - Wikipedia taxonomy
worrms - WoRMS API client
pegax - parse taxonomic names
Taxonstand - Plant List access (*)
R taxonomy task view:
taxize
taxize
access to more than 20 data sources
consistent interfaces for tasks:
- search
- taxonomic ids
- hierarchy
- up/down-stream
- children
- synonyms
- name resolution
taxize citations
with > 50 citations, some e.g.'s
- Retrieving taxa names from large biodiversity data collections using a flexible matching workflow
- Comparative genomics in the Asteraceae reveals little evidence for parallel evolutionary change in invasive taxa
- Are all species necessary to reveal ecologically important patterns?
- Long-lasting effects of land use history on soil fungal communities in second-growth tropical rain forests
- Taxonomic similarity, more than contact opportunity, explains novel plant-pathogen associations between native and alien taxa
- Diversity of ecosystems: Variation in network structure among food webs.
- camtrapR: an R package for efficient camera trap data management
- Genome size variation and species diversity in salamander families
representative taxize use case
"retrieve classification hierarchy from the Integrated Taxonomic Information System"
taxa
taxa
taxonomic classes for R: taxon IDs, names, ranks, data sources
taxonomic classes w/ & w/o data
manipulate taxonomic classes: grow, prune, combine, calculate, etc.
manipulate taxonomic classes while maintaining link to data
taxa as foundation software
idea is to integrate taxa in other software, e.g.:
in development ver. taxize (see taxa-ids branch)
Thoughts on taxa?
Would love to know what people think?
pegax
PEG - Parsing Expression Grammar
define rules (e.g., capture any digit)
combine rules to form a grammar
apply grammar to strings (e.g., taxonomic names)
gnparser exists
tdwg17 talk
thanks dmitry

but ...
R and Java don't play nice
R and C++ do play nice
I'm learning from gnparser to implement R/C++ parser
pegax status
not done yet, but
can instead and into the future use Globalnames Resolver as a web service via taxize
taxize::gnr_resolve
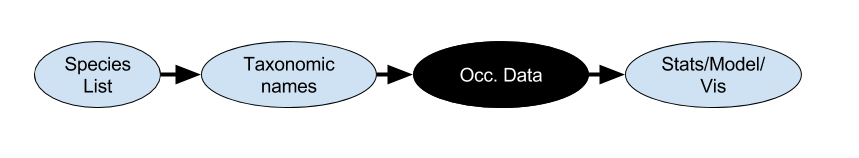
the R biodiversity landscape
R packages
* not rOpenSci
R packages
* not rOpenSci
scrubr - occ. data cleaning
speciesgeocodeR - occ. data cleaning
...
dismo - lots of biodiv. analysis pkgs
GBIF
pre-print in PeerJ: 10.7287/peerj.preprints.3304v1
R: rgbif
Python: pygbif
Ruby: gbifrb
R/Python more for researchers,
Python/Ruby more for web dev
rgbif
> 35 citations, some e.g.'s
Niche dynamics of alien species do not differ among sexual and apomictic flowering plants
Climatological correlates of seed size in Amazonian forest trees
Evolutionary signals of symbiotic persistence in the legume–rhizobia mutualism
Global assessment of arbuscular mycorrhizal fungus diversity reveals very low endemism
Final Report of the Task Group on GBIF Data Fitness for Use in Agrobiodiversity
Plant Distribution Data Show Broader Climatic Limits than Expert-Based Climatic Tolerance Estimates
representative rgbif use case
"... species occurrence data were gathered for major marine phyla ... from geo-referenced specimen data on [GBIF] ... using a polygon search area in ... rgbif"
