Open and reproducible research with R (and web scraping!)
Scott Chamberlain
UC Berkeley / rOpenSci

scotttalks.info/uofo17
LICENSE: CC-BY 4.0
open science/research
open research is badly needed
Retractions

research should be reproducible!
but doing for real is another issue
100 psychology studies
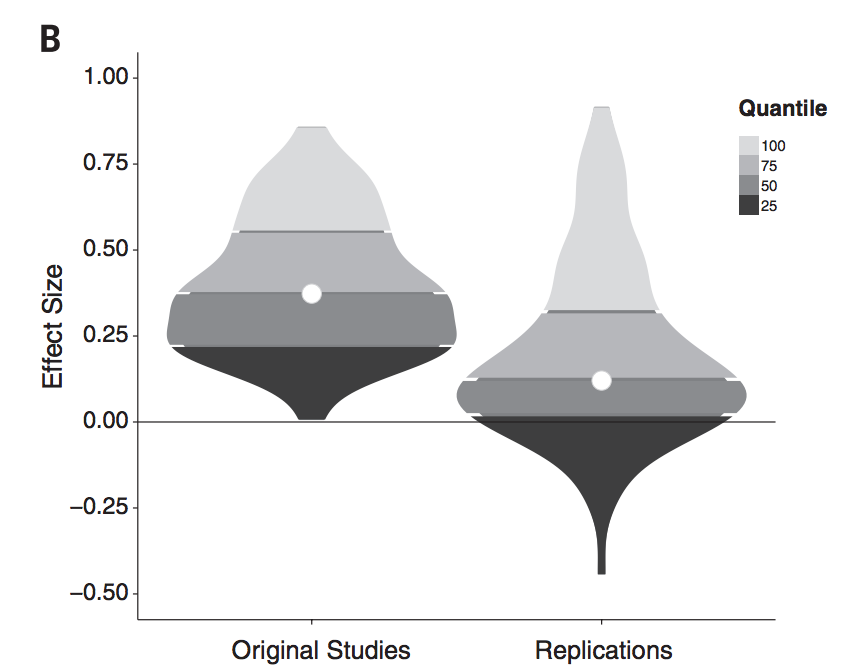
Emergent findings
open data can make a new finding possible
Barriers
Technical
Cultural
Barriers
Technical
Cultural
Cultural barriers
Lack of incentives (carrots)
Lack of pressure (sticks)
Getting scooped ()
Takes too much time! ()
Open science as a lego set

Open research as a lego set
open research may be hard to do
but - you can work on different components
and - individual components are useful on their own
you don't need to do it all at once
Open research components
Open Data
make your data open
funders/journals often requiring this anyway
future self will thank you
Open Data: Venues
- Include data with publications
- Data specific repositories
- Code sharing sites: e.g., GitHub
- so-called Institutional Repositories (IRs) (UO's Scholars Bank)
Open Access
make your papers open
funders often requiring this anyway
talk to your librarians!
Open Access: Preprints
Preprints increasingly allowed by publishers
++ preprint outlets
SocArXiv, PsyArXiv
talk to your librarians!
Open Access: Green OA
Allowed to put up your "authors copy" on your website/etc.
the internet will surface it
Versioning
Versioning

source
Versioning
Including basically all research components:
- Code
- Data
- Metadata
- Text: manuscripts
Why use Versioning?
- failure proofs your work
- allows you to experiment freely!
- Metadata
- Text: manuscripts
Versioning: Git
Resources
- Happy Git with R
- Take a Software Carpentry course
- canonical link git source code and manual
Do all work programatically
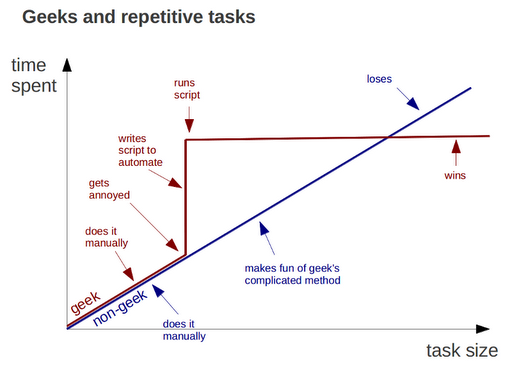
from geeksaresexy.net/2012/01/05/geeks-vs-non-geeks-picture
Do all work programatically
Key to reproduciblity:
Most important person that wants to reproduce your work is you!
Do all work programatically
you and yourself
- one week from now
- two months from now
- & so on
Do all work programatically
allows others to:
- contribute to your work
- check your work
- build on top of your work
research programming languages
research programming languages
are:
the canvas on which to do research
important research programming languages



R language
Open/Rep. Research w/ R
What's the most important thing about R wrt open/reproducible?
R itself -> you're programming!
Open research ecosytsem
rOpenSci does:
the research workflow
Data acquisition
data manipulation/analysis/viz
writing
publish
the research workflow
Data acquisition
data manipulation/analysis/viz
writing
publish
the research workflow
Data acquisition
data manipulation/analysis/viz
writing
publish
the research workflow
Data acquisition
data manipulation/analysis/viz
writing
publish
the research workflow
Data acquisition
data manipulation/analysis/viz
writing
publish
Wrap Up
Open research is essential
Open research tools are useful on their own
rOpenSci: one of the tool makers
Challenges going forward
Largely cultural - will slowly change
Wrap Up
rOpenSci is a community project
Let us know what you need
Help us make better tools
Questions?
let's switch gears ...
Web Data
Web Data: Types
Scraping html
Download files
HTTP
FTP
etc.
APIs
going down: increasing organization and longevity
going up: increasing complexity for user (mostly)
Web Data: Use Cases
Scraping: specialized, one time problems
Download: when you want all the data
APIs: stable, medium sized data
Approaching a website wrt data
Look at menu, header, footer
Look for key words: "API", "Data", "Developers", etc.
Contact a technical (or anyone) person
Browser developer tools are your friends
Scraping as a last resort
Scraping: brief intro
n, def: extract useful bits out of a pile of (typically) html (me, today)
see Wikipedia's def.
Scraping Exercise
navigate to: http://www.goducks.com/cumestats.aspx?path=wsoc
- Install and load
rvestlibrary - Fetch the contents of the URL above
- Pull out the results table
url <- "http://www.goducks.com/cumestats.aspx?path=wsoc"
x <- xml2::read_html(url)
rvest::html_table(x)[[6]]
Scraping: beware
Look on "terms" or related page for any legalese
Probably all good when simply exploring
Take precautions when doing lots of scraping
Look around: StackOverflow, just googling, talk to friends, etc.
Downloading files: intro
(includes web scraping, have to download page to scrape it)
Can be easy - sometimes hard
Protocols: HTTP, FTP, etc. (mostly HTTP)
Authentication sometimes
File sizes can be very large
Does UO have a firewall?
Downloading files in R
lots of ways to download files
many pkgs/fxns for certain file types also download the file for you
recommendation: curl::curl_download - replaces
download.filein base Rthink about where you're putting files!
Downloading Exercise - 1
the file: file
- Download the file using
curl::curl_download - Read the file in however you like
url <- "https://raw.githubusercontent.com/sckott/soylocs/gh-pages/data.csv"
x <- curl::curl_download(url, (f <- tempfile(fileext = ".csv")))
readr::read_csv(x)
Downloading files: misc. topics
Compressed files
Asynchronous downloads can help - check out crul's async vignette
Downloading Exercise - 2
Compressed files
US Dept. Education Academic Libraries Survey 2012
- Download the
.zipfile Academic Libraries Survey 2002 - Uncompress the file
- Read in the
.txtfile
Downloading Exercise - 2
Compressed files
Answer
url <- "https://nces.ed.gov/pubs2006/data/ALS2002_ASCII.zip"
x <- curl::curl_download(url, (f <- tempfile(fileext = ".zip")))
mydir <- file.path(tempdir(), "mydir")
unzip(f, exdir = mydir)
readr::read_tsv(list.files(mydir, full.names=TRUE))
APIs!
API = Application Programming Interface
Rules about how machines/software talk to one another
Can be an API for: Android phone, Linux operating system, an R package, a web service
APIs for data on the web
Web APIs
A server in cloud
Database w/ data
Rules about how to talk to that database
Standardized APIs?
most APIs follow no standard, though loosely follow REST
but some standardized frameworks
Lack of Standardization in APIs
Makes Consuming them Tough
APIs Exercise - 1
Data source: https://usedgov.github.io/api/crdc.html
Make a request for data - any request
goal: make sure everything's working as expected
APIs Exercise - 2
Data source: https://usedgov.github.io/api/crdc.html
- Make a request for data
- Parse data to a list with
resourcesas adata.frame - Pull out all the data for the
TOT_ENR_FandTOT_ENR_Mfields
APIs Exercise - 3
Data source: https://usedgov.github.io/api/crdc.html
- Make a request for data w/ headers
- Get the request headers
- Get the response headers
- Pick a response header and google it/wikipedia it, find out what it is
APIs Exercise - 4
Data source: https://httpbin.org/
- Look up curl options (see
curl::curl_options()), check a few out to see what they do - Make a few requests for data w/ a different curl option specified for each
Look for API Wrappers
Don't reinvent the wheel
Check CRAN, GitHub, etc.
Resources: software
HTTP R pkgs: curl, crul, httr
Scraping pkgs: xml2, rvest, RSelenium
Curated software discussion: Web Technologies TaskView, Open Data TaskView
useR 2016 Workshop: Extracting Data from the Web